Department X-Ray Microscopy
User Service: Cryo-SXT for Life Sciences
Transmission X-ray Tomography for Biological Applications
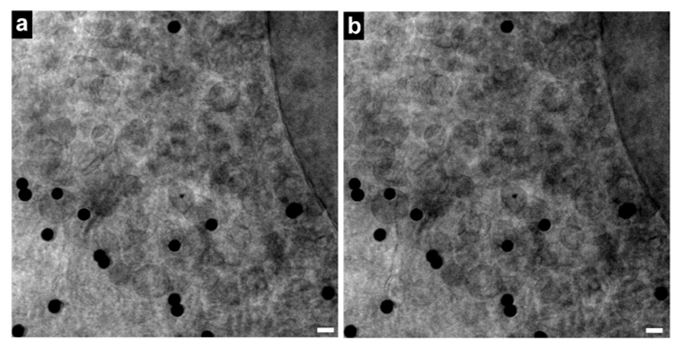
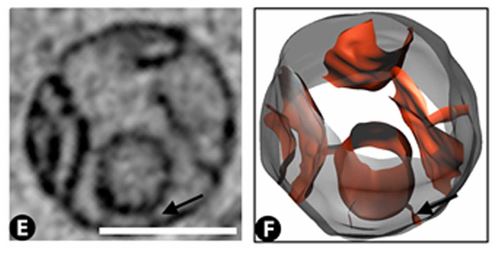
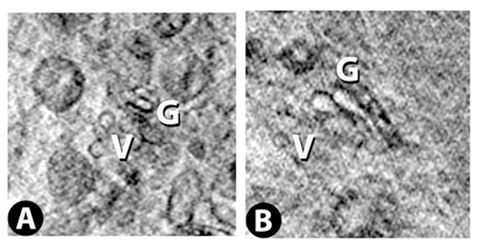
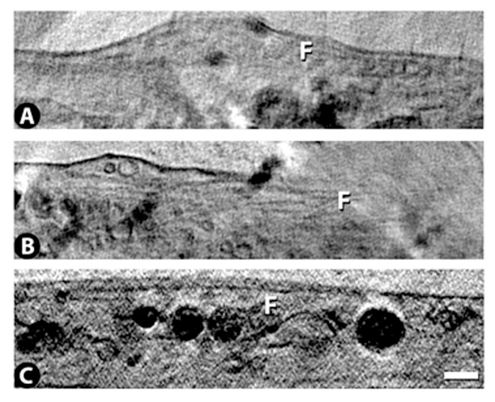
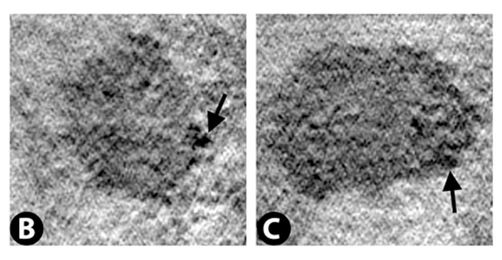
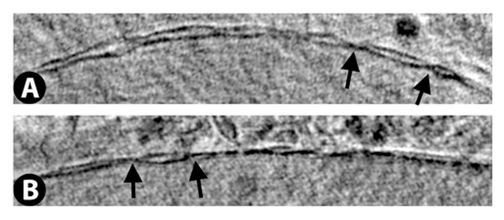
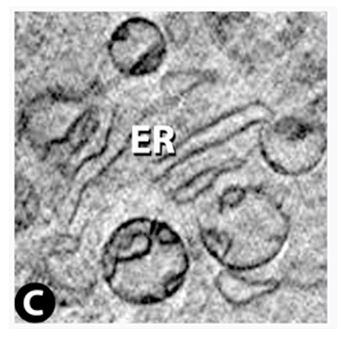
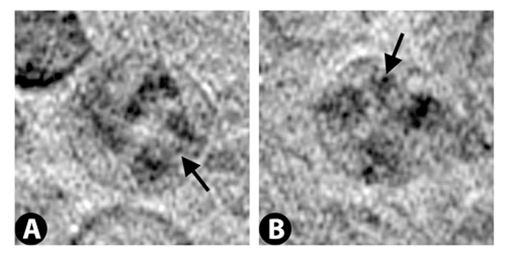
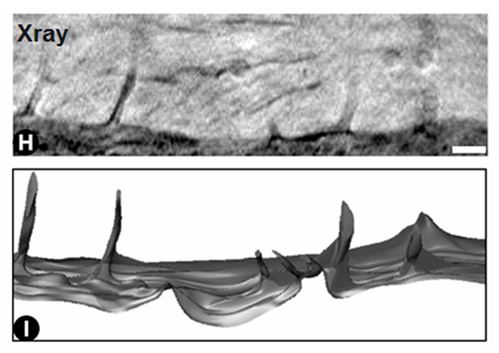
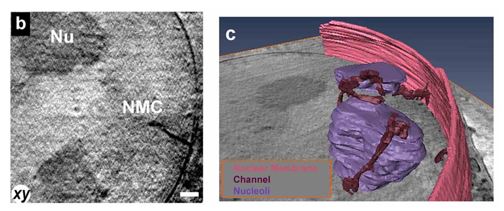
Cryo-soft X-ray tomography can visualize cellular ultrastructure at 30-40 nm resolution in 3D without extensive sample processing such as staining or sectioning. This is accomplished by exploiting the inherent chemical contrast in the spectral region called "water window" (2-4 nm wavelength). In that range, carbon-dense structures (e.g. biomembranes, dense protein assemblies) absorb the soft X-rays much more than the surrounding, less carbon-dense medium (e.g. cytoplasm).
By rotating the sample by defined tilt angles, multiple tilt images can be acquired and used to calculate the 3D ultrastructure of inside the field of view, similarly to cryo-ET. However, much unlike cryo-ET, the thickness of the sample is not limited to a few hundred nanometers. The maximum thickness for biological specimen in our microscope is around 12 - 15 µm, depending on the absorption properties of the specimen.
The ultrastructural information gained by cryo-SXT can also be overlaid with fluorescence microscopy information. For this purpose, our X-ray microscope is also equipped with a cryo-epifluorescence microscope with a green and red channel.
The imaging is performed under cryo-conditions, which protects the sample against irradiation damage and distortions during the acquisition under vacuum.
Vero E6 Tomogram
7.04 sKey Facts for Users
Imaging Parameters:
- Resolution: 30-40 nm in XY
- Field of View per dataset: 12-15 x 12-15 µm
- Energy: For biological specimen, we typically use 510 eV.
- Maximum sample thickness: 12-15 µm. Most adherent mammalian cell lines fit comfortably in this range.
- Tilt range: +-60° to 65°
- Acquisition time: Depends a lot on your specimen, but generally, a full tilt-series acquisition takes typically 20-30 min.
Sample Requirements:
- Chemical fixation: For standard applications, you can use it, but do not have to. It is only necessary if you want to use biohazardous specimen.
- Staining: For cryo-SXT, you do not need to stain your sample at all. It is possible to perform fluorescence imaging of your specimen before cryo-SXT (for instance for correlative studies).
- Specimen support: We typically use standard 3 mm TEM grids with carbon support film. Adherent cell lines can be grown directly on these grids. Suspension cells can be pipetted onto the grids before freezing.
- How to prepare and handle cryo-samples: Typically, plunge freezing is used to vitrify samples. If you do not have a possibility to prepare cryo-samples, we can still assist you. Please get in touch with us.
Microscope Access:
- Proposal writing: Access to the microscope is on a proposal-basis via GATE. There are two calls for proposals per year. In a short 2-3 page proposal, you explain your project and why you want to use cryo-SXT specifically. We are happy to support you with proposal writing.
- Experiment scheduling: Once your proposal has been allocated beamtime, you can coordinate with our beamline staff when you can access the microscope.
- Microscope operation: You do not need to know how to operate the microscope. Our beamline staff will assist you with the measurements.
- Remote acess: Please contact our beamline staff for more information.
Contact and Access
Access to the instrumentation is via the GATE portal on a proposal-basis. It contains further information on current calls for proposals, user access etc. .
If you consider using cryo-SXT for your life science projects and applying for beamtime, please contact us!
Responsible Contacts:
Selected Publications of cryo-SXT in Life Sciences
At a first glance:
- Wüstner, D.; Dupont Juhl, A.; Egebjerg, J.M.; Werner, S.; McNally, J.; Schneider, G.: Kinetic modelling of sterol transport between plasma membrane and endo-lysosomes based on quantitative fluorescence and X-ray imaging data. Frontiers in Cell and Developmental Biology 11 (2023), p. 44936/1-18 DOI: 10.3389/fcell.2023.1144936
- Dyhr, M.C.A.; Sadeghi, M.; Moynova, R.; Knappe, C.; Kepsutlu Çakmak, B.; Werner, S.; Schneider, G.; McNally, J.; Noé, F.; Ewers, H.: 3D surface reconstruction of cellular cryo-soft X-ray microscopy tomograms using semisupervised deep learning. PNAS 120 (2023), p. e2209938120/1-10, DOI: 10.1073/pnas.2209938120
- Kepsutlu, B., Wycisk, V., Achazi, K., Kapishnikov, S., Pérez-Berná, A.J., Guttmann, P., Cossmer, A., Pereiro, E., Ewers, H., Ballauff, B., Schneider, G., McNally, J.G.: Cells Undergo Major Changes in the Quantity of Cytoplasmic Organells after Uptake of Gold Nanoparticles with Biologically Relevant Surface Coatings. ACS Nano 14 (2) (2020), 2248-2264, DOI: 10.1021/acsnano.9b09264
- Review: Groen, J., Conesa J.J., Valcárcel, R., Pereiro, E.: The cellular landscape by cryo soft X-ray tomography. Biophys. Rev. 2019 Jul 4;11(4):611–619. DOI: 10.1007/s12551-019-00567-6
- W.G. Müller, J.B. Heymann, K. Nagashima, P. Guttmann, S. Werner, S. Rehbein, G. Schneider, J.G. McNally: Towards an atlas of mammalian cell ultrastructure by cryo soft X-ray tomography. J. Struct. Biol. 177 (2012), 179-192, DOI: 10.1016/j.jsb.2011.11.025
More Publications (selection)
-
Egebjerg, J.M.; Szomek, M.; Thaysen, K.; Dupont Juhl, A.; Kozakijevic, S.; Werner, S.; Pratsch, C.; Schneider, G.; Kapishnikov, S.; Ekman, A.; Röttger, R.; Wüstner, D.: Automated quantification of vacuole fusion and lipophagy in Saccharomyces cerevisiae from fluorescence and cryo-soft X-ray microscopy data using deep learning. Autophagy 20 (2024), p. 902-922 DOI: 10.1080/15548627.2023.2270378
-
Dupont Juhl, A.; Kozakijevic, S.; Willms, T.; Egebjerg, J.M.; Szomek, M.; Thaysen, K.; Pratsch, C.; Werner, S.; Schneider, G.; Müller, P.; Wüstner, D.: Direct Observation of Uptake and Dissolution of Cholesterol Crystals by Macrophages Using Combined Fluorescence and X-ray Microscopy. Microscopy and Microanalysis 29 (2023), p. 1158-1159 DOI: 10.1093/micmic/ozad067.592
- Spedalieri, C., Szekeres, G.P., Werner, S., Guttmann, P., Kneipp, J.: Probing the Intracellular Bio-Nano Interface in Different Cell Lines with Gold Nanostars. Nanomaterials 11 (2021), 1183 DOI: 10.3390/nano11051183
- Dupont Juhl, A., Lund, F.W., Jensen, M.L.V., Szomek, M., Heegaard, C.W., Guttmann, P., Werner, S., McNally, J., Schneider, G., Kapishnikov, S., Wüstner, D.: Niemann Pick C2 protein enables cholesterol transfer from endo-lysosomes to the plasma membrane for efflux by shedding of extracellular vesicles. Chemistry and Physics of Lipids 235 (2021), 105047 DOI: 10.1016/j.chemphyslip.2020.105047
For a more comprehensive list of publications, see our group's publication list.